High-Speed Protein Movies to Facilitate Drug Design
Researchers from the University of Southampton have developed technology to help scientists observe proteins in motion. Understanding how proteins move will allow novel drugs to be designed.
X-ray crystallography is a scientific method which produces a 3D picture of molecules with exquisite, atomic-level detail. It has been used to determine the structure of many thousands of proteins – nature’s molecular machines.
The new challenge for scientists is to use X-ray crystallography to create ‘movies’ of proteins in action. This is staggeringly difficult, with only a handful made. Efforts are hampered by the protein structure being blurred while capturing each frame in the movie.
Now, a team from the Institute for Life Sciences at the University of Southampton, working in collaboration with Diamond Light Source and Douglas Instruments, have addressed this challenge by miniaturising protein crystals and developing a method for fast mixing. Their findings are published in the journal International Union of Crystallography.
The first author, Jack Stubbs, explains how the crystals are analysed: “The protein crystals are delivered, one by one, into an intense X-ray beam to capture snapshots of the crystals from every possible angle. The resulting scattered, X-ray patterns are used to decipher the protein structure.
“The method can also be used to capture 3D pictures of the proteins at different time-points. Piece these together and you effectively have a movie of proteins in action, which gives clues to their function.”
During this process, it is important that the X-ray ‘snapshots’ capture proteins in the same shape, at the same moment in time, to avoid ‘blurring’ their structure. It is this problem the team has grappled with.
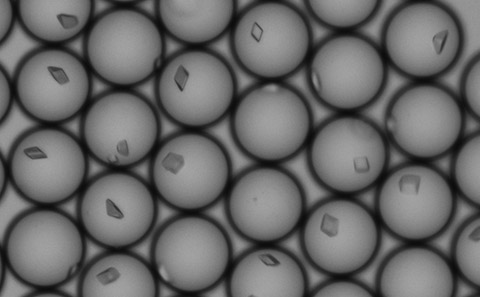
The researchers have developed a ‘droplet microfluidic method’ to produce millions of droplets – each of which acts as a miniature test tube, as small as a trillionth of a litre, for proteins to crystallise. At such vanishingly small volumes, the tiny amount of protein available means that crystals can only grow to a few microns in length – about the same size as a bacterium.
Droplets also have a peculiar ability for mixing. As a droplet moves, the contents circulate, much like stirring, to drive rapid mixing. The team demonstrated mixing times approaching 1 millisecond, 1/1000th of a second. This sets a new standard for time-resolved crystallography.
Lead scientist, Dr Jonathan West, expressed his excitement: “Our research marks a significant advance for time-resolved serial crystallography. The ability to engineer microcrystal size and start reactions by rapid mixing opens up exciting possibilities for understanding protein structural dynamics with millisecond precision.”
The implications of this research extend beyond the laboratory. By sharing these cutting-edge methods with crystallography scientists, the researchers aim to greatly extend our understanding of how proteins move, how motion lends itself to function and how drugs can alter movement.
The research was conducted at the Macromolecular Crystallisation Facility within the School of Biological Sciences at the University of Southampton.