Accelerating Drug Discovery: University of Bremen Introduces GlycoSHIELD for Faster Results
The proteins on the surface of our body’s cells are coated with sugars that influence how cells interact with their environment and with pathogens. They therefore play an important role in the research and production of medicines. The newly developed, environmentally friendly and open source software toolkit GlycoSHIELD now enables fast and efficient analyzes of protein sugar shields. The new approach was developed as part of the German-Polish Dioscuri program of the Max Planck Society in collaboration with the University of Bremen, the Inserm research facility in France and the Academia Sinica in Taiwan.
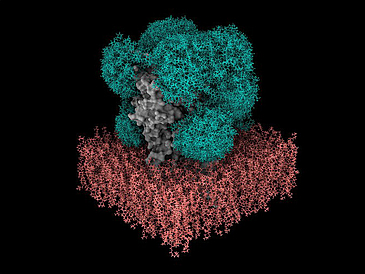
In order to understand how proteins work and how they influence the development and progression of diseases, researchers need to know their three-dimensional atomic structure. In addition to experimental methods, they also use computer simulations. The artificial intelligence a lphafold can, for example, reliably predict the spatial structure of a protein from the sequence of the individual protein building blocks, the amino acids. However, over 75% of the proteins present on the surface of our cells not only consist of amino acid building blocks, but are also covered by sugars, so-called glycans. These form very dynamic “shields” around the proteins. It is currently not known exactly how these protective shields behave and how they influence the binding of drug molecules. Due to their mobility and variability, predicting the morphology of sugars, i.e. the shape and structure of the sugar molecules, has previously been difficult and resource-intensive. Dr. Isabell Louise Grothaus from the Hybrid Materials Interfaces Group led by Professor Lucio Colombi Ciacchi at the University of Bremen has been working on computer simulations of glycans for four years and has developed software called GlyCONFORMER for the analysis and classification of their mobility and variability. Their expertise and software were crucial in the development of the new GlycoSHIELD toolkit, which enables rapid and realistic modeling of the sugar chains on protein surfaces, as just reported in the prestigious journal Cell.
From thousands of hours to just a few minutes
Sugars strongly influence how proteins interact with other molecules. For example, the sugar shield on the spike protein of the SARS-Cov-2 coronavirus makes it difficult for antibodies to recognize the virus, thereby hiding it from our immune system. This shows the relevance of sugar shields for drug development – pharmaceutical research could benefit from routine prediction of their morphology and dynamics. However, until now, predicting the structure of sugar layers using computer simulations was only possible with specialist knowledge and special supercomputers. In many cases this required thousands or even millions of hours of computing. With GlycoSHIELD, the team of scientists led by Dr. Mateusz Sikora, head of the Dioscuri Center for Modeling Post-Translational Modifications at the Jagiellonian University in Krakow, created an environmentally friendly open source alternative. “Our approach reduces the resources, the computing time and the required technical know-how,” says Sikora. “Anyone can now calculate the arrangement and dynamics of sugar molecules on proteins within a few minutes on their PC.” The software could be helpful in the development of drugs or vaccines, for example in immunotherapy against cancer cells, which have a very specific profile Have glycans on their surface.
A puzzle game made of sugar
How did the team go about achieving such a high increase in efficiency? The researchers created and analyzed a library with thousands of likely 3D positions of the most common forms of sugar chains on proteins found in humans and microorganisms. Through long simulations and experiments, they found that it is possible to reliably predict how certain glycans will behave. It is initially not necessary to include the interaction between the shells of cells or parts of the proteins with the attached sugar. The algorithm is based on these findings. “GlycoSHIELD users only need to specify the protein and the locations where the sugars are bound. Our software then puzzles them onto the surface in the most likely arrangement,” explains Sikora. “We were able to reproduce the sugar shields of the spike protein very well, they look exactly as we see them in our experiments!”. With GlycoSHIELD it is now possible to supplement both new and existing protein structures with sugar information. The scientists: inside also used GlycoSHIELD to reveal the pattern of sugars on the GABAA receptor, an important target for sedatives and anesthetics.