
Purdue Researchers Investigate Noninvasive Method for Sampling Drug Response
Harnessing a pervasive type of cellular messenger shows early experimental promise as a routine way of sampling and monitoring the body’s response to prescription drug exposure.
Purdue University-led experiments have successfully isolated drug-metabolizing enzymes from extracellular vesicles (EVs), which are widely secreted throughout the body for cellular communication.
“EVs are involved in almost every kind of activity you can think of,” said Purdue’s W. Andy Tao, professor of biochemistry. These activities include transporting metabolic enzymes produced during drug interactions in the liver.
Tao and his associates have begun developing a patent-pending EV method for detecting proteins involved in drug absorption, distribution, metabolism and excretion (ADME). “If successful, this would be tremendously beneficial, especially for drug development companies,” Tao said.
Tao and 10 co-authors from Purdue and St. Jude Children’s Research Hospital in Memphis, Tennessee, published the details of their method in the Proceedings of the National Academy of Sciences Nexus. The lead authors were Purdue alumnus Xiaofeng Wu and Menchus Quan, a Purdue doctoral student in biological sciences.
According to Tao and his co-authors, the method may also lend itself to monitoring a patient’s drug metabolic performance and how drugs interact with each other. Both are key aspects of personalized medicine.
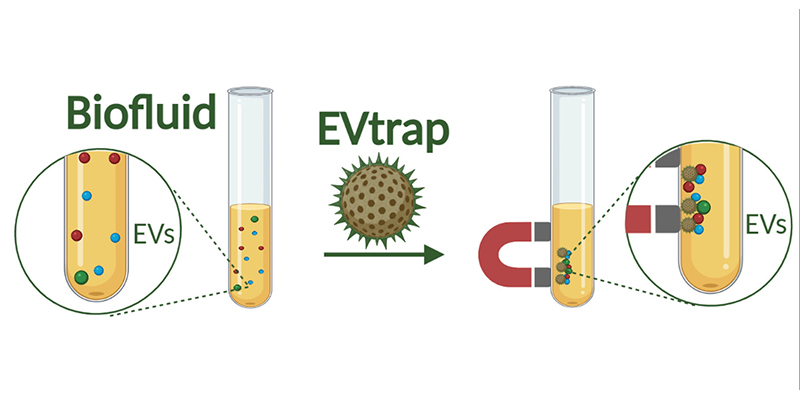 The EVtrap technology uses magnetic beads to rapidly isolate and identify large quantities of proteins from extracellular vesicles, which cells use in their molecular delivery systems. (Image provided by Tymora Analytical Operations) Download image
The EVtrap technology uses magnetic beads to rapidly isolate and identify large quantities of proteins from extracellular vesicles, which cells use in their molecular delivery systems. (Image provided by Tymora Analytical Operations) Download imageThe Centers for Disease Control and Prevention has reported that four in 10 Americans aged 65 or over had been taking at least five prescription drugs as recently as 2018. Half or more of the drugs would be metabolized by the same enzyme, Tao noted. One drug may interfere with another drug through affecting the enzyme’s activity. Such interference can render one drug less effective than another.
Standard practice may dictate that a typical person should take two pills of a given medicine daily. But the response and effectiveness could vary greatly depending on the individual, Tao noted, adding, “And we don’t have a good way to monitor it.”
EVs are already being developed for noninvasive early disease detection. Tao and his associates have contributed to these efforts by developing a technique that may reveal signs of Parkinson’s disease in urine samples.
Involved in that work were three Purdue alumni and co-authors of the PNAS Nexus paper: Wu, now a senior scientist at Pfizer; Marco Hadisurya, a proteomics bioinformatics specialist in biochemistry at Purdue; and Anton Iliuk, president and chief technology officer of Purdue spinoff company Tymora Analytical Operations of West Lafayette.
EVs may help drug discovery and development organizations with federal reporting requirements. The Food and Drug Administration requires the organizations to report how well metabolic enzymes and other proteins involved in ADME respond to their products.
The PNAS Nexus paper notes that “in clinical scenarios, real-time ADME protein detection still lacks effective analytical methods for precise drug efficacy and toxicity evaluation for each patient.”
Liver cells metabolize most drugs, Tao said. A major group of the metabolic enzyme involved in that process are the human cytochrome P450 enzymes. “Almost 80% of our drugs are metabolized by this group of enzymes,” he said.
The researchers tested their ability to detect two metabolic enzymes produced in mice that correspond to similar enzymes in humans after treating them with two widely used drugs. One of the enzymes corresponded to P450 in humans.
One of the tested drugs was rifampicin, an antibiotic well known for inducing a strong response from P450 proteins. The other was dasatinib, a leukemia chemotherapy drug that Purdue alumnus and PNAS Nexus paper co-author Jun Yang studies in his laboratory at St. Jude Children’s Research Hospital.
Analyzing EVs from blood samples for drug metabolism presents challenges because the EVs the samples contain could have come from many different organs. The researchers would prefer to see EVs that were mostly secreted from liver cells, but those account for only a small percentage of blood samples.
“This is an extremely challenging issue and very noisy if you use that as the source for the measurement. Sensitivity is key,” Tao said.
The team applied two separate methods to detect liver EVs with more sensitivity. One was the EV total recovery and purification (EVtrap) technology that Tao co-invented. The technology helps to more efficiently isolate EVs that contain P450 and other metabolic enzymes. The other was a mass spectrometry method that precisely measures subtle variations in the protein composition of tested samples.
“If you don’t have an efficient way to isolate EVs and an efficient way to measure these P450 metabolic enzymes in the EVs, you don’t get a result,” Tao said. “It’s understandable why this has not been addressed in the pharmaceutical field.”
Tao and his associates managed, however, to quantify 34 proteins involved in the ADME process. They found elevated levels of two proteins in mouse plasma EVs that have human counterparts, a response to daily rifampicin dosages. To a lesser extent, the researchers also recorded elevated levels of one of those proteins in response to dasatinib.
“The strategy demonstrated the feasibility of discovering ADME markers for specific drug monitoring by liquid biopsy,” the authors reported in their PNAS Nexus paper. The National Institutes of Health partly funded this research. Tao disclosed his innovation to the Purdue Innovates Office of Technology Commercialization, which has applied for a patent on the intellectual property. Industry partners interested in further developing the innovation should contact Joe Kasper, [email protected], about track code 69103.
